Biological Systems Modeling
Biological Systems
Biomedical research has generated a vast amount of experimental data that can reveal reaction pathways, molecular transport, and population dynamics. Along with experimental data, there is also a growing repository of models that can be used to interpret and refine understanding of biological systems. One collection of models is built on the System Biology Markup Language (SBML), including hundreds of contributions. Many models in this collection have detailed reaction metabolic pathways that describe biological systems, including cause and effect relationships in the human body. While simulations of these biological systems have been successfully applied for many years, the alignment to available measurements continues to be a challenge. Researchers have reported that best available solution techniques continue to limit the size of the reconciliation of model and measurements to small and medium size problems.
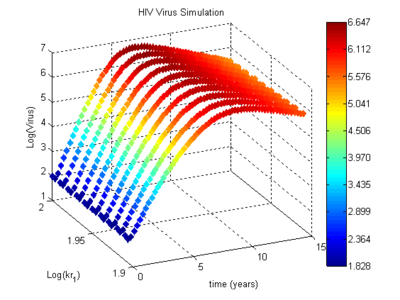
Large-scale estimation solution techniques have recently been applied in refining and chemical process applications. The aim this project is to apply the most advanced solution techniques from the hydrocarbon processing industry to the systems biology field. An interface between SBML and APMonitor, a cutting-edge software package used by petrochemical companies for estimation with large-scale models of differential and algebraic (DAE) systems. Support for this research is given by Vertex Pharmaceuticals.
References
- Lewis, N.R., Hedengren, J.D., Haseltine, E.L., Hybrid Dynamic Optimization Methods for Systems Biology with Efficient Sensitivities, Special Issue on Algorithms and Applications in Dynamic Optimization, Processes, 2015, 3(3), 701-729; doi:10.3390/pr3030701. Article
- Safdarnejad, S.M., Hedengren, J.D., Lewis, N.R., Haseltine, E., Initialization Strategies for Optimization of Dynamic Systems, Computers and Chemical Engineering, 2015, Vol. 78, pp. 39-50, DOI: 10.1016/j.compchemeng.2015.04.016. Article
